I’ve been dabbling with Machine Learning Procedures for Neo4j for a little bit now. Besides providing machine learning capabilities for your graph data, one goal was to support storage, visualization and querying of machine learning models (neural network configurations) in the graph database.
I tried to import TensorFlow models via their protobuf serialization, but unfortunately, the examples protobuf files I found, were incompatible with their Java TensorFlow Protobuf library (maven). If any of you has any insights there I would be supper happy to make that work.
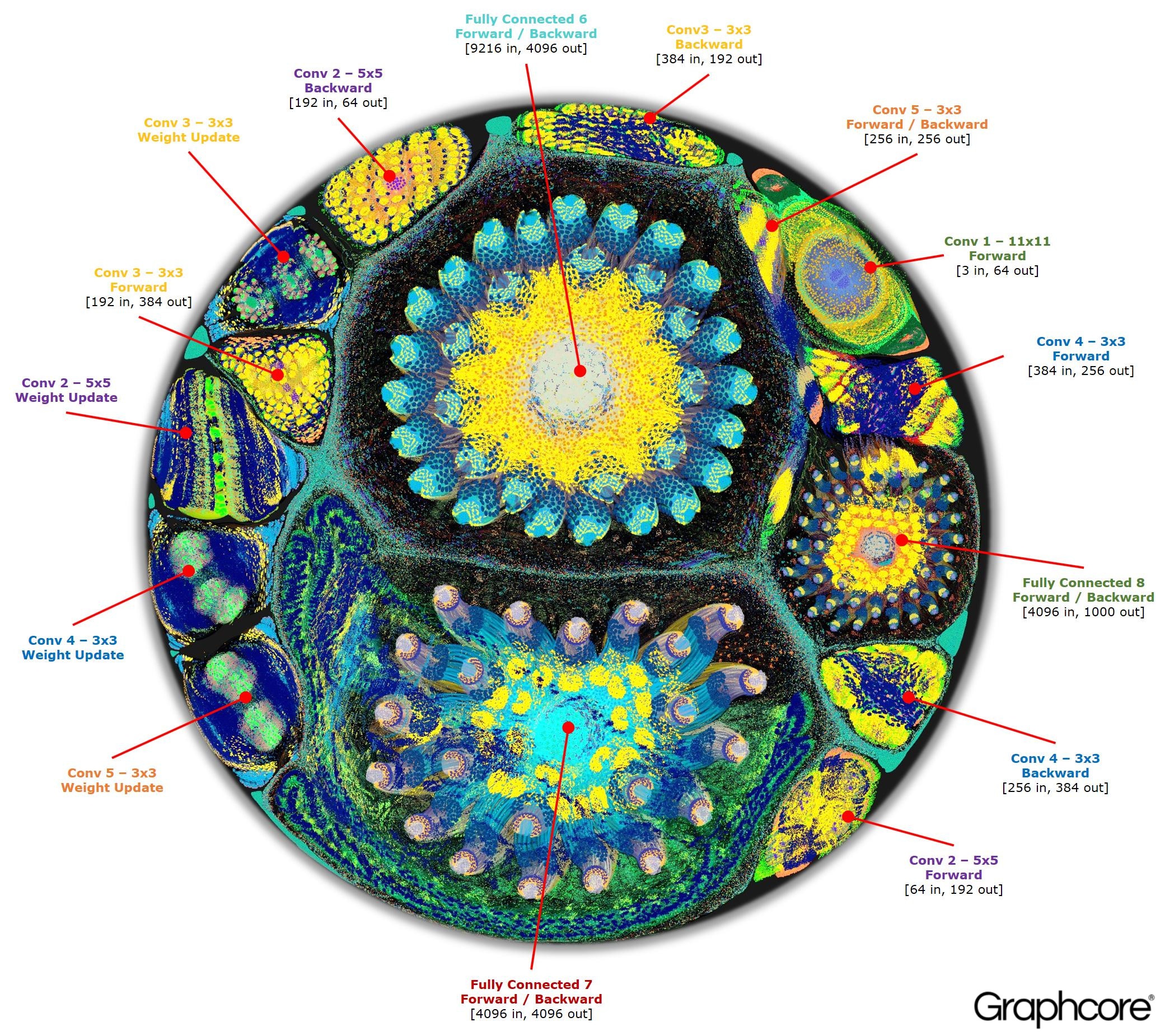
I was inspired to look into putting neural network models into the graph by this amazing article from graphcore.ai which features impressive visualizations of compute networks and a really interesting discussion of the topic.
Last week, my colleague Andreas pointed me to the recent announcment of Facebook and Microsoft to open-source the ONNX "Open Neural Network Exchange" project for an interoperable AI/ML/DeepLearning format.
Microsoft and Facebook have announced a new open source project today that’s aimed at creating a shared model representation for neural networks across different programming frameworks. Called the Open Neural Network Exchange (ONNX), the new project will make it possible to share models across the Cognitive Toolkit, PyTorch, and Caffe2.
I found the ONNX project on GitHub and while it was quite sparsely documented, I saw a Juypter notebook, containing a few examples for small network descriptions. So I thought, this could be a worthwhile candidate for my stated goal. Let’s spend our Saturday morning getting this to work.
Installing ONNX
Unfortunately there was some serious Yak-shaving involved for me, to install ONNX. So perhaps reading about my mishaps is helpful for others.
I tried to follow the instructions in the README but they didn’t really work.
First of all you have to have a protoc compiler on your system, otherwise it complains during installation.
So I figured out where to get it from and ran brew install protobuf which worked.
Then I tried to install ONNX via pip but that failed.
Someone later commented that one shouldn’t do that in the root of the checked out repository.
mh$ pip install onnx
Collecting onnx
Downloading onnx-0.1.tar.gz (588kB)
100% |████████████████████████████████| 593kB 1.3MB/s
...
Successfully built onnx
Installing collected packages: onnx
Successfully installed onnx-0.1
python -c 'import onnx'
Traceback (most recent call last):
File "<string>", line 1, in <module>
File "onnx/__init__.py", line 7, in <module>
from . import checker, helper
File "onnx/checker.py", line 14, in <module>
from onnx import defs
File "onnx/defs/__init__.py", line 6, in <module>
import onnx.onnx_cpp2py_export as C
Back then I couldn’t figure out why and created an issue on their repository though, which gathered some discussion since.
Then I tried to install it from source, which also failed and made me install first pytest and then pybind11 from source.
That also didn’t work that well (failing in some tests) but I somehow could get it installed and then the manual install of ONNX succeeded.
Later, in the issue I createdm I was pointed to the recursive submodule for pybind11
So you have to get the ONNX source, either with git clone --recursive https://github.com/onnx/onnx or do a git submodule update --init after cloning.
brew install protobuf
git clone https://github.com/onnx/onnx
cd onnx
git submodule update --init
Submodule 'third_party/pybind11' (https://github.com/pybind/pybind11.git) registered for path 'third_party/pybind11'
Cloning into '/Users/mh/d/python/onnx/third_party/pybind11'...
Submodule path 'third_party/pybind11': checked out 'a1041190c8b8ff0cd9e2f0752248ad5e3789ea0c'
pip install -e .
Obtaining file:///Users/mh/d/python/onnx
Requirement already satisfied: six in /usr/local/lib/python2.7/site-packages (from onnx==0.1)
Requirement already satisfied: numpy in /usr/local/lib/python2.7/site-packages (from onnx==0.1)
Requirement already satisfied: protobuf in /usr/local/lib/python2.7/site-packages (from onnx==0.1)
Requirement already satisfied: setuptools in /usr/local/lib/python2.7/site-packages (from protobuf->onnx==0.1)
Installing collected packages: onnx
Found existing installation: onnx 0.1
Uninstalling onnx-0.1:
Successfully uninstalled onnx-0.1
Running setup.py develop for onnx
Successfully installed onnx
python -c 'import onnx'
Phew, that worked (for me on Saturday, it was 2 hours later).
ONNX Examples
Now let’s look at the examples in the Juypter Notebook.
They are pretty straightforward, so I only looked at the last one, which set up a complete graph.
from onnx.onnx_pb2 import *
from onnx import checker, helper
# An node that is also a graph
graph = helper.make_graph(
[
helper.make_node("FC", ["X", "W1", "B1"], ["H1"]),
helper.make_node("Relu", ["H1"], ["R1"]),
helper.make_node("FC", ["R1", "W2", "B2"], ["Y"]),
],
"MLP",
["X", "W1", "B1", "W2", "B2"],
["Y"]
)
print(str(graph))(It didn’t find the registry import for me, so I left it off.)
Here is the structure.
node {
input: "X"
input: "W1"
input: "B1"
output: "H1"
op_type: "FC"
}
node {
input: "H1"
output: "R1"
op_type: "Relu"
}
node {
input: "R1"
input: "W2"
input: "B2"
output: "Y"
op_type: "FC"
}
name: "MLP"
input: "X"
input: "W1"
input: "B1"
input: "W2"
input: "B2"
output: "Y"
ir_version: 1
Please note that these repeated input, output and node attributes (note, singular!) are actually a kind of collection, we get back to that soon.
So we have a graph with a bunch of nodes, which have inputs and outputs pointing from or to other nodes. Sounds perfect for our needs.
Quick Neo4j Intro
In a graph database like Neo4j you store your connected data as Nodes and Relationships, both with labels and arbitrary attributes. In that way our conceptional model is the same as your object model, is the same as your graph model, and your visualization.
The core essence of that graph model is patterns where nodes are connected to each other via relationships. You can use these patterns on a whiteboard, diagram, or visualization but also as ascii-art to find and create them in the graph query language Cypher (think "SQL for graphs").
Each node is surrounded by parentheses like a circle (:Person {name:Michael}) and
relationships form ascii-art arrows: (me)-[:LOVES]->(you).
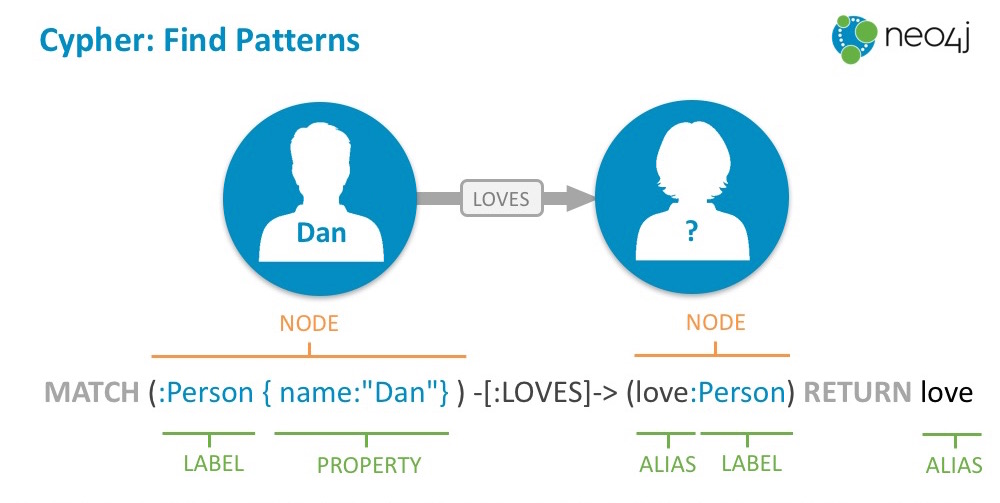
You would send data from ONNX to Neo4j by using the Python driver and sending parameterized Cypher statements to the database. You can find the full driver API docs here.
You can run Neo4j via Docker (docker run -p 7474:7474 -p 7687:7687 neo4j) on the Neo4j Sandbox or on any cloud platform.
Just grab the relevant auth information (after setting the initial password) and database URL from your installation.
pip install neo4j-driver
from neo4j.v1 import GraphDatabase, basic_auth
driver = GraphDatabase.driver("bolt://localhost:7687", auth=basic_auth("neo4j", "test"))
session = driver.session()
# create a pattern, 2 nodes one relationship
statement = """
CREATE (p:Person {name:$name})-[:USES]->(d:Database {name:$what})
"""
# parameters to avoid injection
params = {"name":"Michael","what":"Neo4j"}
session.run(statement,params).consume
# query for a pattern, "what does a person use"
result = session.run(
"MATCH (who:Person)-[:USES]->(tech) RETURN who.name as name, tech.name as what")
# output result columns
for record in result:
print("%s %s" % (record["name"], record["what"]))
session.close()Onnx2Graph
Ok, it shouldn’t be so hard to import that graph object into neo4j, we just need to turn this data into a property graph model.
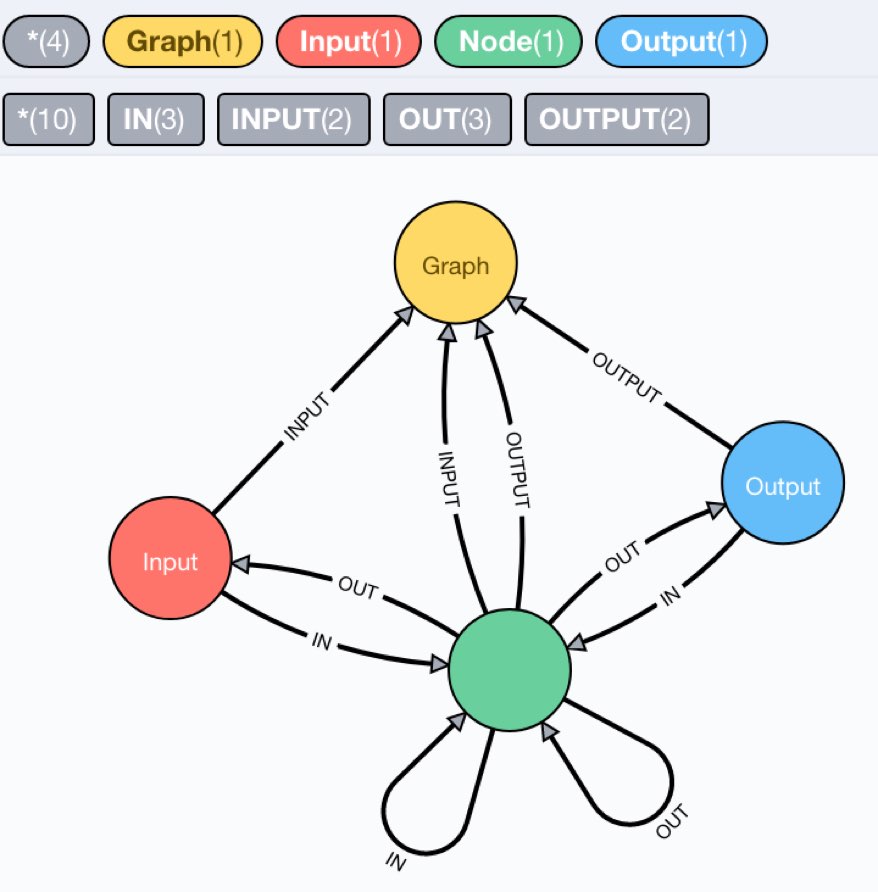
We have a node representing the compute graph, labeled :Graph and pointing with INPUT/OUTPUT relationships to the inputs and outputs of the computation.
Then inside the computation we have :Node labeled neurons, which get and send information via IN and OUT relationships to their respective inputs and outputs.
An example model is here:
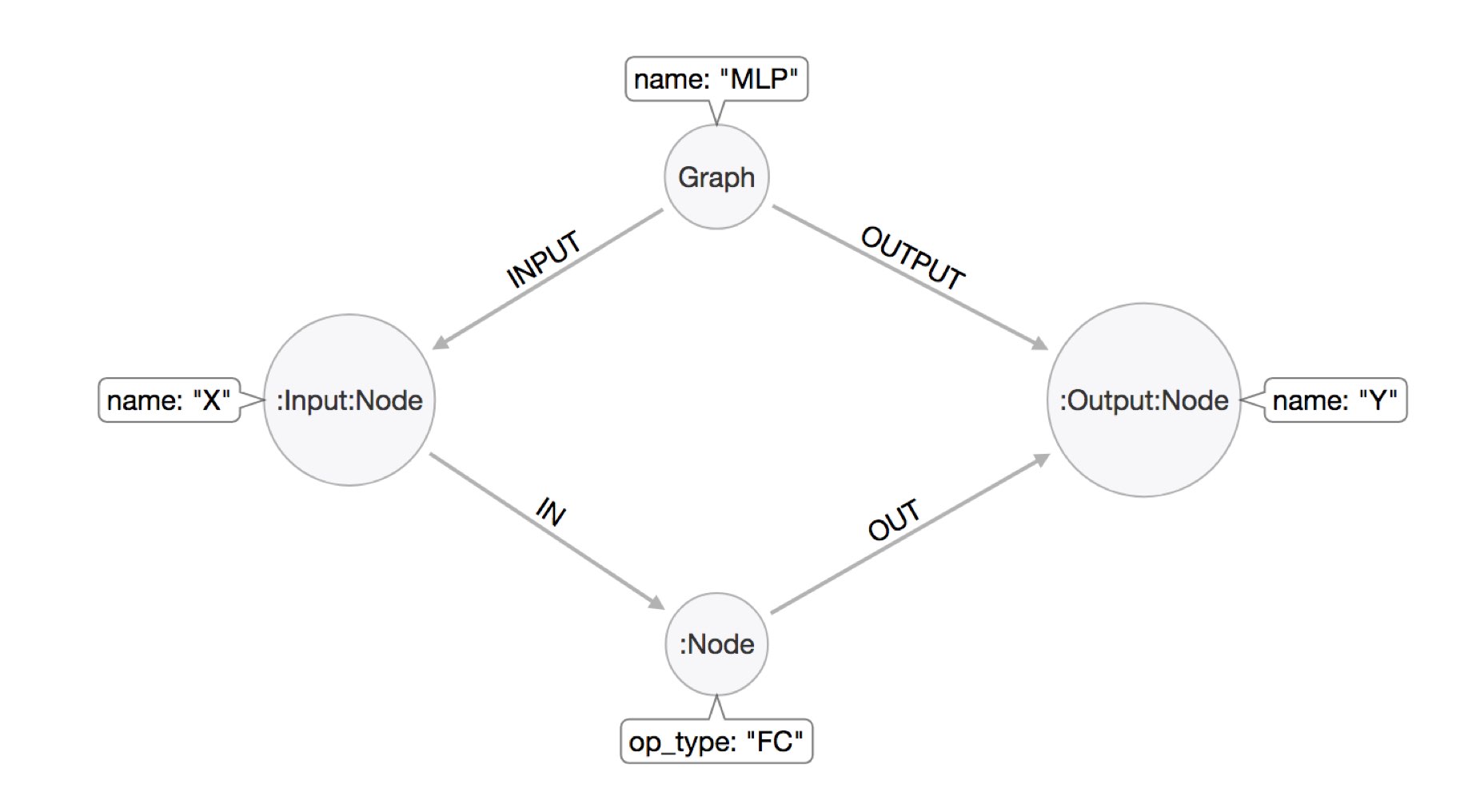
To turn the Python object from ONNX into a Neo4j datastructure, we need a small Cypher statement that takes the whole object, iterates over ites parts and turns it into our graph model.
That annotated statement looks like this ($graph is the parameter we pass in):
// create the :Graph node
MERGE (g:Graph {name:$graph.name}) SET g += $graph.attr
// create input nodes in the context of this graph and connect them
FOREACH (name in $graph.inputs | MERGE (i:Node {name:name})-[:INPUT]->(g) SET i:Input)
// create output nodes in the context of this graph and connect them
FOREACH (name in $graph.outputs | MERGE (g)<-[:OUTPUT]-(o:Node {name:name}) SET o:Output)
WITH *
// iterate over all nodes of the graph
UNWIND $graph.nodes as node
// create the compute node, set it's attributes (attributes can vary widely)
CREATE (n:Node) SET n += node.attr
// find-or-create input nodes to our node and connect them, mark them as input nodes
FOREACH (name in node.inputs | MERGE (i:Node {name:name}) SET i:Input MERGE (i)-[:IN]->(n))
// find-or-create output nodes to our node and connect them, mark them as output nodes
FOREACH (name in node.outputs | MERGE (o:Node {name:name}) SET o:Output MERGE (n)-[:OUT]->(o));Now we only have to turn the ONNX object in a proper nested dictionary, that represented the graph structured we wanted to pass into our statement.
I didn’t realize first, that I have to convert the nested protobuf collections manually.
My initial attempt looked like this, and "succeeded", and looked ok in Python.
params = {"graph":{"name":graph.name, "attr":{"ir_version":graph.ir_version}, "inputs":graph.input, "outputs":graph.output,
"nodes": map(lambda n : {"attr":{"op_type":n.op_type},"inputs":n.input,"outputs":n.output}, graph.node)}}
{'graph': {'inputs': [u'X', u'W1', u'B1', u'W2', u'B2'],
'nodes': [{'inputs': [u'X', u'W1', u'B1'], 'attr': {'op_type': u'FC'}, 'outputs': [u'H1']},
{'inputs': [u'H1'], 'attr': {'op_type': u'Relu'}, 'outputs': [u'R1']},
{'inputs': [u'R1', u'W2', u'B2'], 'attr': {'op_type': u'FC'}, 'outputs': [u'Y']}],
'name': u'MLP', 'outputs': [u'Y'], 'attr': {'ir_version': 1L}}}When I tried to insert that data into my Neo4j instance, though it failed with error:
ValueError: Values of type <class 'google.protobuf.internal.containers.RepeatedScalarFieldContainer'> are not supported
So initially I transferred all 4 of those sneaky RepeatedScalarFieldContainer’s manually with lambdas `map(lambda x: str(x), graph.input) which was not really pretty.
Googling for RepeatedScalarFieldContainer I found some API docs, which pointed me to the _values field in the superclass, which I could use instead.
params = {"graph":{"name":graph.name, "attr":{"ir_version":graph.ir_version}, "inputs":graph.input._values, "outputs":graph.output._values,
"nodes": map(lambda n : {"attr":{"op_type":n.op_type},"inputs":n.input._values,"outputs":n.output._values}, graph.node)}}So, now we could put it all together.
from neo4j.v1 import GraphDatabase, basic_auth
driver = GraphDatabase.driver("bolt://localhost:7687", auth=basic_auth("neo4j", "test"))
session = driver.session()
statement = """
MERGE (g:Graph {name:$graph.name}) SET g += $graph.attr
FOREACH (name in $graph.inputs | MERGE (i:Node {name:name})-[:INPUT]->(g) SET i:Input)
FOREACH (name in $graph.outputs | MERGE (g)<-[:OUTPUT]-(o:Node {name:name}) SET o:Output)
WITH *
UNWIND $graph.nodes as node
CREATE (n:Node) SET n += node.attr
FOREACH (name in node.inputs | MERGE (i:Node {name:name}) SET i:Input MERGE (i)-[:IN]->(n))
FOREACH (name in node.outputs | MERGE (o:Node {name:name}) SET o:Output MERGE (n)-[:OUT]->(o));
"""
params = {"graph":{"name":graph.name, "attr":{"ir_version":graph.ir_version}, "inputs":graph.input._values, "outputs":graph.output._values,
"nodes": map(lambda n : {"attr":{"op_type":n.op_type},"inputs":n.input._values,"outputs":n.output._values}, graph.node)}}
session.run(statement,params).consume
session.close()I put all the code also into this [Jupyter Notebook: onnx2graph^].
After running this statement, we can visit the Neo4j browser and see our tiny compute graph in its full beauty.
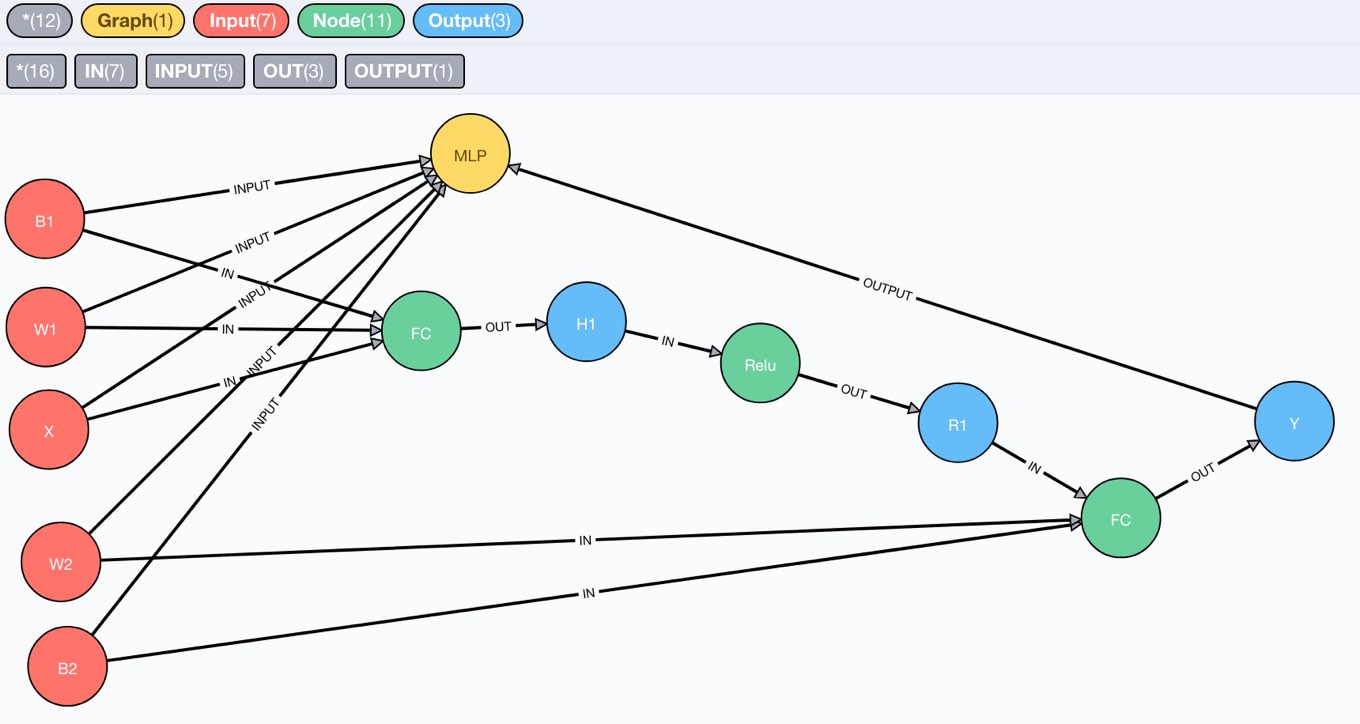
We can also inspect the schema, which is what we expected.
After this initial achievement, at the end of the weekend, I looked at the ONNX support for torch
The torch.onnx module contains functions to export models into the ONNX IR format. These models can be loaded with the ONNX library and then converted to models which run on other deep learning frameworks.
With this example, which sounded exciting, starting from my original graphcore.ai inspiration:
Example: End-to-end AlexNet from PyTorch to Caffe2 Here is a simple script which exports a pretrained AlexNet as defined in torchvision into ONNX. It runs a single round of inference and then saves the resulting traced model to alexnet.proto
Let’s import the full AlexNet ONNX file:
If you like this approach, please let me know and I can turn this into a proper library.
As soon as there are Java bindings for ONNX I can add support for this to the neo4j-ml-procedures
Have fun